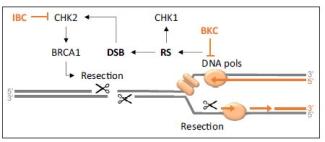
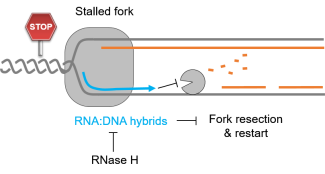
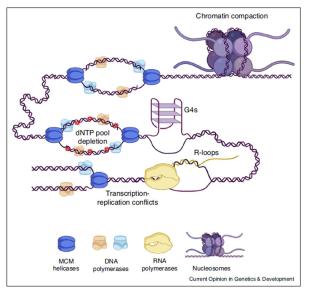
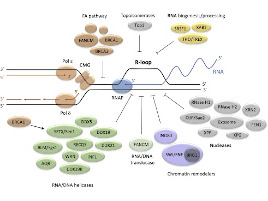
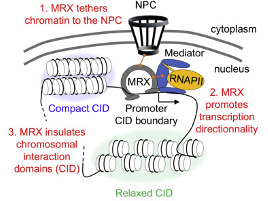
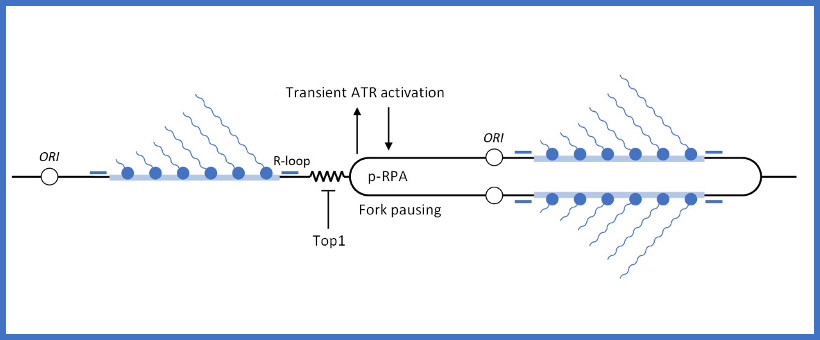
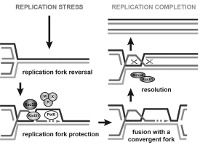
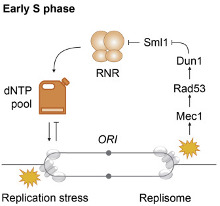
The human body is made of 3.7x1013 cells, which contain about two meters of DNA each. As our cells undergo a total of 1016 divisions in a life time, they synthesize therefore more than 2x1016 meters of DNA, which represents 130,000 times the distance from Earth to the sun! This daunting task is executed by micro-machines called replisomes, containing hundreds of proteins. Replisomes assemble at replication origins and generate DNA structures called replication forks. As they progress along the chromosomes, replisomes often encounter obstacles such as DNA lesions or transcription complexes, leading to replication fork arrest. Stalled forks are fragile structures that can give rise to chromosome breaks and trigger genomic instability if they are not rapidly restarted. Fork stalling occurs even more frequently in cancer cells due to the deregulation of oncogenic pathways. Oncogene-induced replication stress (RS) promotes genomic instability and is therefore the driving force of tumorigenesis. However, RS represents also the Achilles’ heel of cancer cells as it interferes with cell proliferation and sensitizes them to chemotherapeutic agents. Understanding how normal and cancer cells respond to replication stress represents therefore a major challenge in cancer biology.
Our group investigates the cellular responses to replication stress in budding yeast and in human cell lines. Owing to the small size of its genome and the power of molecular genetics, budding yeast is an invaluable model organism to study the RS response and to characterize novel mechanisms that are conserved in human cells and are relevant to cancer biology. This is achieved through the use of powerful technologies to monitor the progression, arrest and recovery of replication forks. These methods include single-molecule approaches such as DNA combing and DNA fiber spreading, chromosome-based assays such as pulsed-field gel electrophoresis and NGS-based assays such as ChIP-seq, BrdU-IP-seq, DRIP-seq and BLESS. Together, these methods provide a comprehensive view of the replication stress response in yeast and human cells, from individual DNA molecules to whole genomes.
To further investigate the links between replication stress and cancer, we have recently teamed up with the group of Jérôme Moreaux (Hematology Department of the University Hospital of Montpellier) who is an expert of the pathophysiology of malignant plasma cells and in particular of Multiple Myeloma (MM).