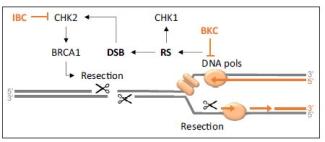
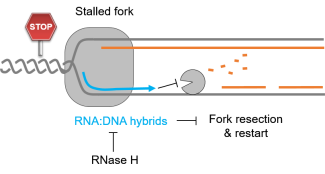
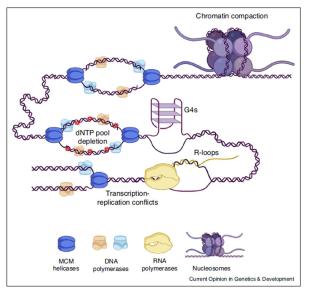
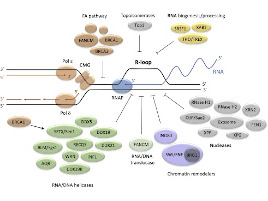
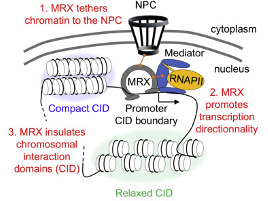
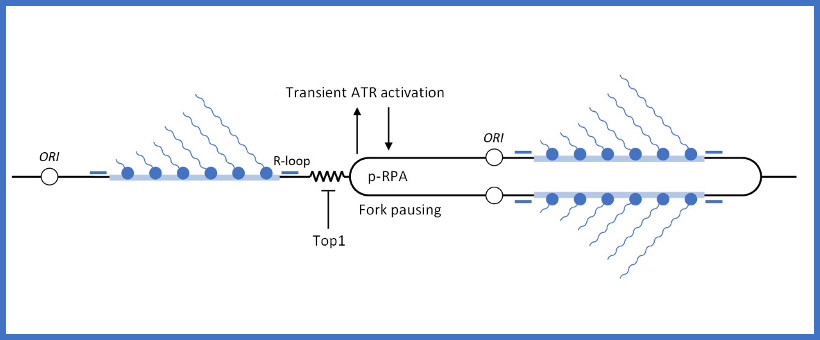
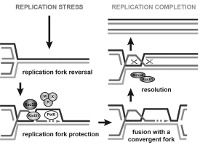
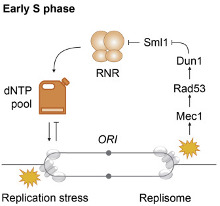
Le corps humain est constitué de 3.7x1013 cellules, contenant chacune environ deux mètres d’ADN. Sachant que nos cellules font un total de 1016 cycles de division cellulaire au cours de notre existence, elles synthétisent donc plus de 2x1016 m d’ADN, ce qui représente 130 000 fois la distance de la terre au soleil! Cette tâche considérable est assurée par des micro-machines appelées réplisomes, contenant plusieurs centaines de protéines. Les réplisomes sont assemblés au niveau des origines de réplication et génèrent des structures d’ADN appelées fourches de réplication. Au cours de leur déplacement le long des chromosomes, les réplisomes rencontrent souvent des obstacles tels que des lésions de l’ADN ou des complexes de transcription, entrainant un arrêt des fourches de réplication. Les fourches bloquées sont des structures fragiles, pouvant induire la formation de cassures de l’ADN et une instabilité génétique si elles ne sont pas rapidement redémarrées. Le blocage des fourches est encore plus fréquents dans les cellules tumorales suite à la dérégulation de voies oncogéniques, ce qui provoque un stress réplicatif. Ce stress réplicatif induit par les oncogènes joue un rôle central dans la tumorigenèse en induisant de l’instabilité génétique. Cependant, il représente aussi le talon d’Achille de la tumeur car il ralentit la prolifération des cellules et les rend plus sensibles à la chimiothérapie. Comprendre comment les cellules normales et tumorales répondent au stress réplicatif représente donc un enjeu majeur en cancérologie.
Notre équipe étudie les mécanismes de réponse au stress réplicatif chez la levure et dans des lignées de cellules humaines. Compte tenu de la petite taille de son génome et de la puissance des outils de génétique moléculaire, la levure S. cerevisiae est un modèle de choix pour caractériser de nouveaux mécanismes de réponse au stress et étudier ensuite leur rôle dans les cellules tumorales. Pour ce faire, nous utilisons des technologies de pointe permettant de suivre la progression, l’arrêt et le redémarrage des fourches. Il s’agit notamment d’approches ‘molécule unique’ comme le peignage moléculaire, de techniques permettant d’étudier la structure des chromosomes par électrophorèse en champs pulsés et d’approches pan-génomiques (séquençage à haut débit), comme le ChIP-seq, le BrdU-IP-seq, le DRIP-seq et le BLESS. L’utilisation combinée de ces différentes approches permet d’avoir une vision globale de la réponse au stress réplicatif chez la levure et l’Homme, de la molécule d’ADN unique au génome entier.
Afin d’explorer l’impact potentiel de ces nouvelles connaissances pour le traitement du cancer, nous nous sommes récemment associés à l’équipe de Jérôme Moreaux (Service d’Hématologie du CHU de Montpellier), qui est un expert reconnu dans le domaine des hémopathies malignes et notamment du Myélome Multiple.