Chromatine et Biologie Cellulaire

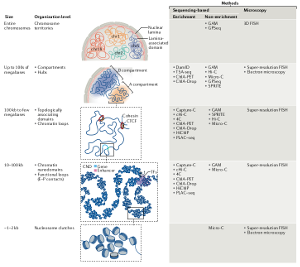
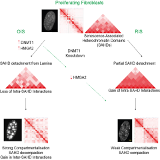
Le génome est plus qu'une suite linéaire de gènes. Il est compacté dans l’espace tridimensionnel du noyau cellulaire en structures tertiaires et quaternaires d'ordre supérieur. Les gènes flanquants s'organisent dans des domaines chromosomiques qui sont caractérisés par de types différents de chromatine, actifs ou répressifs.
Dans l'espace 3D, chaque site à l’intérieur d’un domaine contacte les autres sites à l’intérieur du domaine plus fréquemment que les sites d’autres domaines.
Cette propriété permet de définir les domaines physiques ou topologiquement associés (TAD). Au niveau du chromosome, les TAD individuels forment des contacts avec d'autres TAD, préférentiellement du même type, afin de construire des architectures 3D ordonnées appelées territoires chromosomiques. Enfin, différents chromosomes s'organisent de manière non aléatoire dans l'espace nucléaire.
Par conséquent, le génome eucaryote est hautement organisé en 3D et cette régulation peut être transmise ou modulée pendant la vie des cellules et des organismes. Cette chorégraphie chromosomique sophistiquée implique des milliers de différents acteurs - séquences d'ADN, ARNs et protéines - mais plutôt que de se combiner en un nombre infini de formes, ces composantes organisent un nombre relativement limité de types de chromatine, soit actifs, soit répressifs.
En particulier, deux groupes principaux de facteurs régulateurs du génome sont constitués par les protéines Polycomb (PcG) et Trithorax (trxG). Les protéines PcG maintiennent la mémoire des états silencieux de l'expression de gènes à travers la physiologie cellulaire et les divisions cellulaires multiples, tandis que les membres du trxG maintiennent des états actifs de la chromatine.
Ces protéines sont capables de reconnaître les états chromatiniens de leurs gènes cibles et de maintenir ces états au fil des divisions cellulaires même après la disparition des régulateurs transcriptionnels qui les ont induits en premier lieu. Remarquablement, ces états peuvent également être transmis à une fraction de la descendance au cours de plusieurs générations.
Dans notre laboratoire, nous visons à comprendre le principe régissant l'organisation du génome en 3D, ses implications fonctionnelles et les mécanismes moléculaires par lesquels les protéines PcG et trxG régulent leurs gènes cibles, transmettent l'héritage des états chromatiniens et orchestrent le développement.
Pour atteindre cet objectif, nous utilisons une variété d'approches et de techniques complémentaires dans les domaines de la biologie moléculaire, cellulaire et de développement, de la génomique et de la bioinformatique.
Publications de l'équipe
Phase separation and inheritance of repressive chromatin domains.
Akilli N, Cheutin T, Cavalli G
Drosophila TET acts with PRC1 to activate gene expression independently of its catalytic activity.
Gilbert G, Renaud Y, Teste C, Anglaret N, Bertrand R, Hoehn S, Jurkowski TP, Schuettengruber B, Cavalli G, Waltzer L, Vandel L

Transient loss of Polycomb components induces an epigenetic cancer fate.
Parreno V, Loubiere V, Schuettengruber B, Fritsch L, Rawal CC, Erokhin M, Győrffy B, Normanno D, Di Stefano M, Moreaux J, Butova NL, Chiolo I, Chetverina D, Martinez AM, Cavalli G
Epigenetic inheritance in adaptive evolution.
Sabarís G, Fitz-James MH, Cavalli G
Repression and 3D-restructuring resolves regulatory conflicts in evolutionarily rearranged genomes.
Ringel AR, Szabo Q, Chiariello AM, Chudzik K, Schöpflin R, Rothe P, Mattei AL, Zehnder T, Harnett D, Laupert V, Bianco S, Hetzel S, Glaser J, Phan MHQ, Schindler M, Ibrahim DM, Paliou C, Esposito A, Prada-Medina CA, Haas SA, Giere P, Vingron M, Wittler L, Meissner A, Nicodemi M, Cavalli G, Bantignies F, Mundlos S, Robson MI
A shared ancient enhancer element differentially regulates the bric-a-brac tandem gene duplicates in the developing Drosophila leg.
Bourbon HG, Benetah MH, Guillou E, Mojica-Vazquez LH, Baanannou A, Bernat-Fabre S, Loubiere V, Bantignies F, Cavalli G, Boube M
Comprehensive characterization of the epigenetic landscape in Multiple Myeloma.
Alaterre E, Ovejero S, Herviou L, de Boussac H, Papadopoulos G, Kulis M, Boireau S, Robert N, Requirand G, Bruyer A, Cartron G, Vincent L, Martinez AM, Martin-Subero JI, Cavalli G, Moreaux J
HiCmapTools: a tool to access HiC contact maps.
Chang JM, Weng YF, Chang WT, Lin FA, Cavalli G
SETDB1/NSD-dependent H3K9me3/H3K36me3 dual heterochromatin maintains gene expression profiles by bookmarking poised enhancers.
Barral A, Pozo G, Ducrot L, Papadopoulos GL, Sauzet S, Oldfield AJ, Cavalli G, Déjardin J
Mechanisms of Polycomb group protein function in cancer.
Parreno V, Martinez AM, Cavalli G
Molecular mechanisms of transgenerational epigenetic inheritance.
Fitz-James MH, Cavalli G
Clinical Correlations of Polycomb Repressive Complex 2 in Different Tumor Types.
Erokhin M, Chetverina O, Győrffy B, Tatarskiy VV, Mogila V, Shtil AA, Roninson IB, Moreaux J, Georgiev P, Cavalli G, Chetverina D
Understanding 3D genome organization by multidisciplinary methods
Ivana Jerkovic´ & Giacomo Cavalli
test
test1, test2
Regulation of single-cell genome organization into TADs and chromatin nanodomains
Szabo Q., Donjon A., Jerkovic I., Papadopoulos G.L., Cheutin T., Bonev B., Nora E., Bruneau B.G., Bantignies F., Cavalli, G.
4D Genome Rewiring during Oncogene-Induced and Replicative Senescence
Sati S., Bonev, B., Szabo, Q., Jost, D., Bensadoun, P., Serra, F., Loubiere
Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping.
Loubiere V, Papadopoulos GL, Szabo Q, Martinez AM, Cavalli G
+
Chromatine et Biologie Cellulaire

Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping.
Loubiere V, Papadopoulos GL, Szabo Q, Martinez AM, Cavalli G
+
Chromatine et Biologie Cellulaire
Global chromatin conformation differences in the Drosophila dosage compensated chromosome X.
Pal K, Forcato M, Jost D, Sexton T, Vaillant C, Salviato E, Mazza EMC, Lugli E, Cavalli G, Ferrari F
+
Chromatine et Biologie Cellulaire
Global chromatin conformation differences in the Drosophila dosage compensated chromosome X.
Pal K, Forcato M, Jost D, Sexton T, Vaillant C, Salviato E, Mazza EMC, Lugli E, Cavalli G, Ferrari F
+
Chromatine et Biologie Cellulaire
The multiscale effects of polycomb mechanisms on 3D chromatin folding.
Cheutin T, Cavalli G
+
Chromatine et Biologie Cellulaire
The multiscale effects of polycomb mechanisms on 3D chromatin folding.
Cheutin T, Cavalli G
+
Chromatine et Biologie Cellulaire
Higher-Order Chromosomal Structures Mediate Genome Function.
Jerković I, Szabo Q, Bantignies F, Cavalli G
+
Chromatine et Biologie Cellulaire
Higher-Order Chromosomal Structures Mediate Genome Function.
Jerković I, Szabo Q, Bantignies F, Cavalli G
+
Chromatine et Biologie Cellulaire

Advances in epigenetics link genetics to the environment and disease
Giacomo Cavalli and Edith Heard
+
Chromatine et Biologie Cellulaire

Advances in epigenetics link genetics to the environment and disease
Giacomo Cavalli and Edith Heard
+
Chromatine et Biologie Cellulaire
Principles of genome folding into topologically associating domains.
Szabo Q, Bantignies F, Cavalli G
Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms.
Cardozo Gizzi AM, Cattoni DI, Fiche JB, Espinola SM, Gurgo J, Messina O, Houbron C, Ogiyama Y, Papadopoulos GL, Cavalli G, Lagha M, Nollmann M
Cell Fate and Developmental Regulation Dynamics by Polycomb Proteins and 3D Genome Architecture.
Loubiere V, Martinez AM, Cavalli G
EZH2 is overexpressed in transitional preplasmablasts and is involved in human plasma cell differentiation.
Herviou L, Jourdan M, Martinez AM, Cavalli G, Moreaux J
+
Maintien de l'intégrité du génome au cours de la réplication
PRC2 targeting is a therapeutic strategy for EZ score defined high-risk multiple myeloma patients and overcome resistance to IMiDs.
Herviou L, Kassambara A, Boireau S, Robert N, Requirand G, Müller-Tidow C, Vincent L, Seckinger A, Goldschmidt H, Cartron G, Hose D, Cavalli G, Moreaux J
+
Maintien de l'intégrité du génome au cours de la réplication
Challenges and guidelines toward 4D nucleome data and model standards.
Marti-Renom MA, Almouzni G, Bickmore WA, Bystricky K, Cavalli G, Fraser P, Gasser SM, Giorgetti L, Heard E, Nicodemi M, Nollmann M, Orozco M, Pombo A, Torres-Padilla ME
Loss of PRC1 induces higher-order opening of Hox loci independently of transcription during Drosophila embryogenesis.
Cheutin T, Cavalli G
[EZH2 is therapeutic target for personalized treatment in multiple myeloma].
Herviou L, Cavalli G, Moreaux J
+
Maintien de l'intégrité du génome au cours de la réplication
Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development.
Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G
TADs are 3D structural units of higher-order chromosome organization in Drosophila.
Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M, Bantignies F, Cavalli G
Technical Review: A Hitchhiker's Guide to Chromosome Conformation Capture.
Grob S, Cavalli G
Multi-scale 3D genome rewiring during mouse neural development.
Bonev, B., Mendelson Cohen, N., Szabo, Q., Fritsch, L., Papadopoulos, G., Lubling, Y., Xu, X., Hugnot, JP., Tanay, A., Cavalli, G.
Genome Regulation by Polycomb and Trithorax: 70 Years and Counting
Schuettengruber, B., Bourbon, HM., Di Croce, L., Cavalli, G.
Chromosome topology guides the Drosophila Dosage Compensation Complex for target gene activation
Schauer T, Ghavi-Helm Y, Sexton T, Albig C, Regnard C, Cavalli G, Furlong EE, Becker PB.
Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila
Ciabrelli F, Comoglio F, Fellous S, Bonev B, Ninova M, Szabo Q, Xuéreb A, Klopp C, Aravin A, Paro R, Bantignies F, Cavalli G
Three-Dimensional Genome Organization and Function in Drosophila
Schwartz YB, Cavalli G.
Chromosome conformation capture technologies and their impact in understanding genome function
Sati S, Cavalli G.
Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions
Cattoni DI, Gizzi AMC, Georgieva M, Di Stefano M, Valeri A, Chamousset D, Houbron C, Déjardin S, Fiche JB, González I, Chang JM, Sexton T, Marti-Renom MA, Bantignies F, Cavalli G, Nollmann M.
Organization and function of the 3D genome
Bonev B, Cavalli G.
Chromosome Conformation Capture on Chip (4C): Data Processing
Leblanc B, Comet I, Bantignies F, Cavalli G.
Following the Motion of Polycomb Bodies in Living Drosophila Embryos
Cheutin T, Cavalli G.
Coordinate redeployment of PRC1 proteins suppresses tumor formation during Drosophila development
Loubière, V., Delest, A., Thomas, A., Bonev, B., Schuettengruber, B., Sati, S., Martinez AM., Cavalli, G.
Regulation of Genome Architecture and Function by Polycomb Proteins
Entrevan M, Schuettengruber B, Cavalli G
EZH2 in normal hematopoiesis and hematological malignancies
Herviou L, Cavalli G, Cartron G, Klein B, Moreaux J.
+ Maintien de l'intégrité du génome au cours de la réplication
PRC1 proteins orchestrate three-dimensional genome architecture
Cavalli, G.
Enhancer of zeste acts as a major developmental regulator of Ciona intestinalis embryogenesis
Le Goff E, Martinand-Mari C, Martin M, Feuillard J, Boublik Y, Godefroy N, Mangeat P, Baghdiguian S, Cavalli G
Distinct polymer physics principles govern chromatin dynamics in mouse and Drosophila topological domains
Ea V, Sexton T, Gostan T, Herviou L, Baudement MO, Zhang Y, Berlivet S, Le Lay-Taha MN, Cathala G, Lesne A, Victor JM, Fan Y, Cavalli G, Forné T.
Histone H3 Serine 28 Is Essential for Efficient Polycomb-Mediated Gene Repression in Drosophila
Yung PY, Stuetzer A, Fischle W, Martinez AM, Cavalli G
The Role of Chromosome Domains in Shaping the Functional Genome
Sexton, T., Cavalli, G.
Developmental determinants in non-communicable chronic diseases and ageing
Bousquet J, Anto JM, Berkouk K, Gergen P, Pinto Antunes J, Augé P, Camuzat T, Bringer J, Mercier J, Best N, Bourret R, Akdis M, Arshad SH, Bedbrook A, Berr C, Bush A, Cavalli G, Charles MA, Clavel-Chapelon F, Gillman M, Gold DR, Goldberg M, Holloway JW, Iozzo P, Jacquemin S, Jeandel C, Kauffmann F, Keil T, Koppelman GH, Krauss-Etschmann S, Kuh D, Lehmann S, Lodrup Carlsen KC, Maier D, Méchali M, Melén E, Moatti JP, Momas I, Nérin P, Postma DS, Ritchie K, Robine JM, Samolinski B, Siroux V, Slagboom PE, Smit HA, Sunyer J, Valenta R, Van de Perre P, Verdier JM, Vrijheid M, Wickman M, Yiallouros P, Zins M.
Chromatin driven behavior of topologically associating domains
Ciabrelli F, Cavalli G.
Cooperativity, Specificity, and Evolutionary Stability of Polycomb Targeting in Drosophila
Schuettengruber B, Oded Elkayam N, Sexton T, Entrevan M, Stern S, Thomas A, Yaffe E, Parrinello H, Tanay A, Cavalli G.
Topological organization of Drosophila hox genes using DNA fluorescent in situ hybridization
Bantignies, F., Cavalli, G.
Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains
Jost D, Carrivain P, Cavalli G, Vaillant C.
Identification of Regulators of the Three-Dimensional Polycomb Organization by a Microscopy-Based Genome-wide RNAi Screen
Gonzalez I, Mateos-Langerak J, Thomas A, Cheutin T, Cavalli G.
A RING to Rule Them All: RING1 as Silencer and Activator
Cavalli, G.
Polycomb silencing: from linear chromatin domains to 3D chromosome folding
Cheutin T, Cavalli G
Chromosomes: now in 3D!
Cavalli, G.
Three-dimensional genome organization: a lesson from the Polycomb-Group proteins
Bantignies, F.
PRC2 Controls Drosophila Oocyte Cell Fate by Repressing Cell Cycle Genes.
Iovino N, Ciabrelli F, Cavalli G.
The 3D Genome Shapes Up For Pluripotency.
Sexton T, Cavalli G.
Functional implications of genome topology
Cavalli, G., Mistelli, T.
Polycomb domain formation depends on short and long distance regulatory cues.
Schuettengruber B, Cavalli G.
Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization.
Tanay A, Cavalli G.
Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion
Cheutin T, Cavalli G.
Polycomb: a paradigm for genome organization from one to three dimensions.
Delest A, Sexton T, Cavalli G.
Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome.
Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G.
Molecular biology. EZH2 goes solo
Cavalli, G.
Trithorax group proteins: switching genes on and keeping them active.
Schuettengruber B, Martinez AM, Iovino N, Cavalli G.
Rolling ES Cells Down the Waddington Landscape with Oct4 and Sox2
Iovino, N., Cavalli, G.
Polycomb group proteins: repression in 3D.
Bantignies F, Cavalli G.
Editorial Review
Hetzer, M., Cavalli, G
Chromatin folding : from linear chromosomes to the 4D nucleus.
Cheutin, T., Bantignies, F., Leblanc, B., Cavalli, G.
From linear genes to Epigenetic inheritance of three dimensional Epigenomes.
Cavalli, G.
A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber.
Comet, I., Schuettengruber, B., Sexton, T., Cavalli, G
Polycomb-dependent Regulatory Contacts between Distant Hox Loci in Drosophila,
Bantignies, F., Roure, V., Comet, I., Leblanc, B., Schuttengruber, B., Bonnet, J., Tixier, V., Mas, A., Cavalli, G.
The DUBle life of polycomb complexes
Schuettengruber, B., Cavalli, G
Uncovering a tumor-suppressor function for Drosophila polycomb group genes.
Martinez, AM., Cavalli, G
Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice.
Schuettengruber B, Cavalli G.
Genomic interactions: Chromatin loops and gene meeting points in transcriptional regulation
Sexton, T., Bantignies, F., Cavalli, G.
Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling
Martinez, AM., Schuettengruber, B., Sakr, S., Janic, A., Gonzalez, C., and Cavalli, G.
Functional Anatomy of Polycomb and Trithorax Chromatin Landscapes in Drosophila Embryos
Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G.
Epigenetics and the control of multicellularity. Workshop on Chromatin at the Nexus of Cell Division and Differentiation.
Reuter G, Cavalli G.
Chapter 2 polycomb group proteins and long-range gene regulation.
Mateos-Langerak J, Cavalli G
The Role of RNAi and Noncoding RNAs in Polycomb Mediated Control of Gene Expression and Genomic Programming.
Portoso, M, and Cavalli, G.
Idefix insulator activity can be modulated by nearby regulatory elements.
Brasset E, Bantignies F, Court F, Cheresiz S, Conte C, Vaury C.
Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex.
Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V.
Visualizing macromolecules with fluoronanogold: from photon microscopy to electron tomography.
Cheutin, T., Sauvage, C., Tchélidzé, P., O
Chromosome kissing
Cavalli, G.
Genome Regulation by Polycomb and Trithorax Proteins.
Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G.
Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions.
Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T
Polycomb controls the cell fate.
Negre N, Cavalli G.
Mapping the distribution of chromatin proteins by ChIP on chip.
Negre N, Lavrov S, Hennetin J, Bellis M, Cavalli G.
From genetics to epigenetics: the tale of Polycomb group and trithorax group genes.
Grimaud C, Negre N, Cavalli G.
PRE-Mediated Bypass of Two Su(Hw) Insulators Targets PcG Proteins to a Downstream Promoter.
Comet I, Savitskaya E, Schuettengruber B, Negre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G.
Chromosomal Distribution of PcG Proteins during Drosophila Development.
Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G.
The role of polycomb group proteins in cell cycle regulation during development.
Martinez AM, Cavalli G.
Polycomb group-dependent Cyclin A repression in Drosophila.
Martinez AM, Colomb S, Dejardin J, Bantignies F, Cavalli G.
RNAi components are required for nuclear clustering of Polycomb group response elements.
Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G.
Chromatin and epigenetics in development: blending cellular memory with cell fate plasticity.
Cavalli G.
Cellular memory and dynamic regulation of polycomb group proteins.
Bantignies F, Cavalli G.
Epigenetic inheritance of chromatin states mediated by Polycomb- and trithorax group proteins in Drosophila
Déjardin, J., and Cavalli, G.
Recruitment of Drosophila Polycomb Group proteins to chromatin by DSP1
Déjardin, J., Rappailles, A., Cuvier, O., Grimaud, C., Decoville, M., Locker, D., and Cavalli, G.
Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1
Dejardin J, Cavalli G.
The epigenome network of excellence.
Akhtar A, Cavalli G
Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line
Aravin, A. A., Klenov, M. S., Vagin, V. V., Bantignies, F., Cavalli, G., and Gvozdev, V. A.
Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module
Déjardin, J., and Cavalli, G.
Interaction between GAF and Mod(mdg4) proteins promotes insulator bypass in Drosophila
Melnikova, L., Juge, F., Gruzdeva, N., Mazur, A., Cavalli, G., and Georgiev, P.
Combined immunostaining and FISH analysis of polytene chromosomes
Lavrov, S., Déjardin, J., and Cavalli, G.
SNR1 is an essential subunit in a subset of drosophila brm complexes, targeting specific functions during development
Zraly, C. B., Marenda, D. R., Nanchal, R., Cavalli, G., Muchardt, C., and Dingwal, A.K.
Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila
Sun, L. V., Chen, L., Greil, F., Nègre, N., Li, T. R., Cavalli, G., Zhao, H., Van Steensel, B., and White, K.
Inheritance of Polycomb-dependent chromosomal interactions in Drosophila
Bantignies, F., Grimaud, C., Lavrov, S., Gabut, M., and Cavalli, G.
Chromatin as a eukaryotic template of genetic information
Cavalli, G.
The MYST Domain Acetyltransferase Chameau Functions in Epigenetic Mechanisms of Transcriptional Repression
Grienenberger, A., Miotto, B., Sagnier, T., Cavalli, G., Schramke, V., Geli, V., Mariol, M. C., Berenger, H., Graba, Y., and Pradel, J.
Epigenetic Inheritance of active chromatin after removal of the main transactivator
Cavalli, G., Paro, R.
Mapping DNA target sites of chromatin-associated proteins by formaldehyde cross-linking in Drosophila embryos.
Cavalli, G., Orlando, V., and Paro, R.
The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis
11) Cavalli, G., and Paro, R.
Thèses et hdr
Étude du repliement tridimensionnel de la chromatine en domaines topologiques 27/11/2019
Soutenue par Quentin Szabo le 27/11/2019 sous la direction de Frédéric Bantignies et de Giacomo Cavalli
Rôle dynamique du PRC1 au cours du développement normal et de la tumorigenèse chez Drosophila melanogaster 16/11/2018
Soutenue par Vincent Loubière le 16/11/2018 sous la direction de Anne/Marie Martinez et de Giacomo Cavalli
Caractérisation de la diversité des sites de fixation des protéines du groupe Polycomb chez la Drosophile 29/09/2017
par Marianne Entrevan le 29/09/2017 sous la direction de Giacomo Cavalli et de Bernd Schuttengruber
Héritage transgénérationnel stable d'états chromatiniens alternatifs chez Drosophila melanogaster 16/12/2015
Defended by Filippo Ciabrelli on 16/12/2015 under the supervision of Giacomo Cavalli
Histone H3 Serine 28 is essential for efficient Polycomb/mediated gene repression in Drosophila 09/02/2015
Soutenue par Yuk Kwong Yung le 09/02/2015 sous la direction de Giacomo Cavalli
Ciblage dynamique et différentiel des complexes Polycomb au cours du développement de Drosophila melanogaster 30/11/2012
Anna Delest, 30/11/2012
Fonction différentielle des protéines du groupe Polycomb durant le développement de la Drosophile 24/11/2011
Soutenue par Samy Sakr le 24/10/2011
Etude génomique et structurale de la chromatine chez la Drosophile 24/10/2011
Benjamin Leblanc, 23/06/2011
Le noyau cellulaire et la régulation génique par les protéines du groupe Polycomb 28/10/2010
Bernd Stadelmayer 28/10/2010
Rôle des protéines du groupe Polycomb et trithorax dans l'organisation nucléaire des gènes homéotiques chez Drosophila melanogaster 10/12/2008
Virginie Roure 10/12/2008
Etudes moléculaires des interactions fonctionnelles entre la répression dépendante des protéines du groupe Polycomb et les insulateurs de gipsy chez la drosophile 20/11/2007
Itys Comet 20/11/2007
Recherche des Gènes Cibles des Protéines du Groupe Polycomb (PcG) chez la Drosophile 28/11/2005
Nicolas Nègre 28/11/2005
Séquences et facteurs nécessaires à la mémoire épigénétique des états chromatiniens 27/05/2004
Jérôme Déjardin 27/05/2004