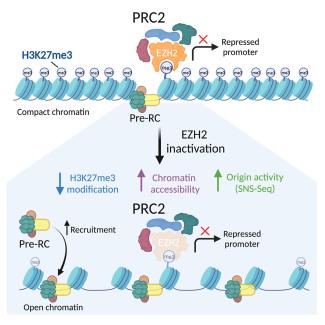
Loss of Ezh2 function remodels the DNA replication initiation landscape
Paulina Prorok, Faezeh Forouzanfar, Nerea Murugarren, Isabelle Peiffer, Romain Charton, Ildem Akerman, Marcel Méchali.
At each division, chromosomes should be duplicated and also maintain the memory of the specific transcription programs that were previously established. Initiation of DNA replication is a precisely regulated process that can start at around 100 000 potential sites dispersed along the genome and are called DNA replication origins (Méchali, 2010; Fragkos et al, 2015). Errors in this process can cause loss or gain of genetic material that will lead to genome instability, a hallmark of cancer cells.
An intriguing and essential aspect of DNA replication origins in mammalian cells is their genetic and epigenetic nature, which remains quite elusive. Their characteristics are generally well established in bacterial, viruses, and unicellular eukaryotes, but unraveling their nature in multicellular eukaryotes has been our mains goal along the years. We also wish to understand how their positions along the genome may play a role in the organization of chromosomes in the nucleus and how they could play an essential role in transcriptional controls during development and cell differentiation.
Our laboratory discovered the first genetic consensus element present at drosophila or mouse replication origins and extended its characterization in human cells. We identified an Origin G-rich Repeated Element (OGRE) that could form G quadruplexes (G4). We also found that this element was orientated 150-300 bp upstream of the initiation site and was nucleosome-free. We could further demonstrate its functional importance in mouse cells. Finally, a striking observation of replication origins in mouse or human cells was their concentration at the borders of TAD domains and their disorganization in cancer cells.
Over the years, we have also discovered and/or characterized several proteins involved in DNA replication, including Cdt1, MCM4 (cdc21), MCM8, MCM9, and Obi1.
Our current research is to link DNA replication origins to the organization of chromatin domains in the nucleus.

Paulina Prorok, Faezeh Forouzanfar, Nerea Murugarren, Isabelle Peiffer, Romain Charton, Ildem Akerman, Marcel Méchali.
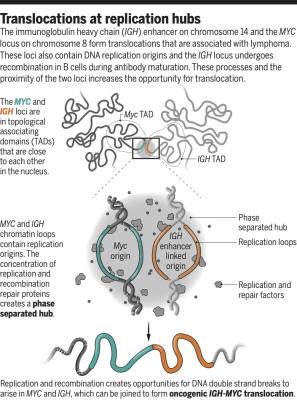
Akerman I, Kasaai B, Bazarova A, Sang PB, Peiffer I, Artufel M, Derelle R, Smith G, Rodriguez-Martinez M, Romano M, Kinet S, Tino P, Theillet C, Taylor N, Ballester B, Méchali M
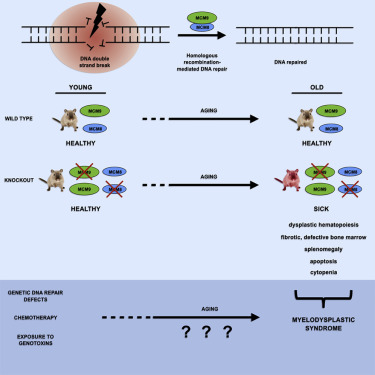
Malik Lutzmann, Florence Bernex, Cindy da Costa de Jesus, Dana Hodroj, Caroline Marty, Isabelle Plo, William Vainchenker, Marie Tosolini, Luc Forichon, Caroline Bret, Sophie Queille, Candice Marchive, Jean-Sébastien Hoffmann, Marcel Méchali

Coulombe P, Nassar J, Peiffer I, Stanojcic S, Sterkers Y, Delamarre A, Bocquet S., Méchali M.
Prorok P., Artufel, M., Aze, A., Coulombe, P., Peifer I., Lacroix L., Guédin A., Mergny J.L., Damaschke J., Schepers A. , Cayrou C, Teulade-Fichou MP, Ballester, B., Méchali M.
Ganier O., Prorok P, Akerman I. , Méchali M.
Aze A, Maiorano D
+
Genome Surveillance and Stability
Aze A, Fragkos M, Bocquet S, Cau J, Méchali M
Brustel J, Kirstein N, Izard F, Grimaud C, Prorok P, Cayrou C, Schotta G, Abdelsamie AF, Déjardin J, Méchali M, Baldacci G, Sardet C, Cadoret JC, Schepers A, Julien E.
+
Biology of Repetitive Sequences
Rodríguez-Martínez, M., Pinzón, N., Ghommidh, C., Beyne, E., Seitz, H., Cayrou, C., Méchali, M.
Bousquet J, Anto JM, Berkouk K, Gergen P, Pinto Antunes J, Augé P, Camuzat T, Bringer J, Mercier J, Best N, Bourret R, Akdis M, Arshad SH, Bedbrook A, Berr C, Bush A, Cavalli G, Charles MA, Clavel-Chapelon F, Gillman M, Gold DR, Goldberg M, Holloway JW, Iozzo P, Jacquemin S, Jeandel C, Kauffmann F, Keil T, Koppelman GH, Krauss-Etschmann S, Kuh D, Lehmann S, Lodrup Carlsen KC, Maier D, Méchali M, Melén E, Moatti JP, Momas I, Nérin P, Postma DS, Ritchie K, Robine JM, Samolinski B, Siroux V, Slagboom PE, Smit HA, Sunyer J, Valenta R, Van de Perre P, Verdier JM, Vrijheid M, Wickman M, Yiallouros P, Zins M.
Hutchins JR, Traver S, Coulombe P, Peiffer I, Kitzmann M, Latreille D, Méchali M.
Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, van Helden J, Méchali M.
Traver S, Coulombe P, Peiffer I, Hutchins JR, Kitzmann M, Latreille D, Méchali M
Fragkos M, Ganier O, Coulombe P, Méchali M
Hutchins JR
Leonard AC, Méchali M
Coulombe, P., Grégoire, D., Tsanov, N., and Méchali, M.
Méchali M, Yoshida K, Coulombe P, Pasero P.
Hua H, Namdar M, Ganier O, Gregan J, Méchali M, Kearsey SE.
Mechali, M.
Cayrou C, Grégoire D, Coulombe P, Danis E, Méchali M
Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, Forichon L, de Massy B, Méchali M.
Estefanía MM, Ganier O, Hernández P, Schvartzman JB, Mechali M, Krimer DB.
Cayrou C, Coulombe P, Puy A, Rialle S, Kaplan N, Segal E, Méchali M.
Ganier O, Bocquet S, Peiffer I, Brochard V, Arnaud P, Puy A, Jouneau A, Feil R, Renard JP, and Méchali M
Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, Méchali M.
Nishiyama A, Frappier L, Méchali M.
Méchali, M.
Cayrou, C., Coulombe, P., Méchali, M
Pillaire, M.J., Selves, J., Gordien, K., Gouraud, P.A., Gentil, C., Danjoux, M., Do, C., Negre, V., Bieth, A., Guimbaud, R., Trouche, D., Pasero, P., Méchali, M., Hoffmann, JS, and Cazaux, C
Méchali, M.
Lutzmann, M., Méchali, M.
Lutzmann M, Méchali M.
Stanojcic, S., Lemaitre, JM., Brodolin, K., Danis, E., Mechali, M.
Cuvier, O., Stanojcic, S., Lemaitre, JM., Méchali, M.
Krasinska, L., Besnard, E., Cot, E., Dohet, C., Méchali, M., Lemaitre, JM., Fisher, D.
Méchali M, and Lutzmann M.
Ganier, O. and Mechali, M.
Auziol C, Mechali M, Maiorano D.
+
Genome Surveillance and Stability
Méchali, M.
Lemaitre, JM., Gregoire, D., Mechali, M.
Grégoire, D., and Méchali, M.
Lutzmann M, Maiorano D, Mechali M.
Maiorano D, Lutzmann M, Mechali M.
Gregoire, D., Brodolin, K., Mechali, M.
Cuvier O, Lutzmann M, Mechali M.
Déjardin, J., Rappailles, A., Cuvier, O., Grimaud, C., Decoville, M., Locker, D., and Cavalli, G.
Lemaitre, J-M., Danis, E., Vassetzky, Y., Pasero, P., and Mechali, M.
Maiorano, D., Cuvier, O., Danis, E., and Mechali, M.
Maiorano, D., Krasinska, L., Lutzmann, M. and Mechali M.
Lutzmann, M., Maiorano, D., and Méchali, M.
Françon, P. ; Lemaitre, JM., Dreyer, C. ; Maiorano, D. ; Cuvier, O. and Marcel Méchali.
Lemaitre, JM., Bocquet, S., Terret, ME., Namdar, M., Ait-Ahmed, O., Kearsey, S., Verlhac, MH., and Méchali, M.
Thepaut, M., Maiorano, D., Guichou, JF., Auge, MT., Dumas, C., Méchali, M., and Padilla, A.
Fisher D., and Marcel Méchali.
Danis, E., Brodolin, K., Menut, S., Maiorano, D., Girard-Reydet, C. and Marcel Méchali.
Girard-Reydet, C., Gregoire, D., Vassetzky, Y., and Marcel Méchali.
Maiorano, D., Rul, W., and Marcel Mechali
Françon, P., and Méchali, M.
Fisher, D., and Mechali, M.
Lemaitre, J-M., and Méchali, M.
Demeret, C., Bocquet, S., Lemaître, J-M., Françon, P., and Méchali, M.
Cohen, S., and Méchali, M.
Lemaître, J-M., Bocquet, S., and Méchali, M.
Méchali, M.
Cohen, S., and Méchali, M.
Demeret, C., Vassetzky, Y. and Mchali, M.
Tada, S., Li, A., Maiorano, D., Méchali, M., and Blow, J.
Maiorano, D., Lemaître, J.M. and Méchali, M.
Vassetzky, Y., Lemaitre, J.M. and Méchali, M.
Fisher, D.L., Mandart, E. and Dorée, M.
Vassetzky, Y., Hair, A., and Méchali M.
Maiorano, D., Moreau, J., and Méchali, M.
Vassetzky, Y.S., Tchang, F., Fanning, E. and Méchali, M.
Tchang, F. and Méchali, M.
Moreau, J., Lebreton, S., Iouzalen, N. and Méchali, M.
Menut, S., Lemaitre, J.M., Hair, A. and Méchali, M.
Françon, P., Maiorano, D. and Méchali, M.
Cohen, S., Menut, S. and Méchali, M.
Lemaitre, J.M., Géraud, G. and Méchali, M.
Hair, A., Prioleau, M.N., Vassetzki, Y. and Méchali, M.
Coué, M., Amariglio, F., Maiorano, D., Bocquet, S. and Méchali, M.
Defended by Joelle Nassar on 25-10-2019
