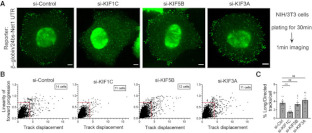
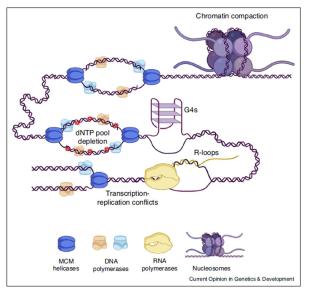
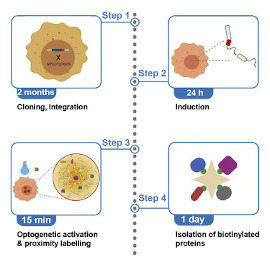
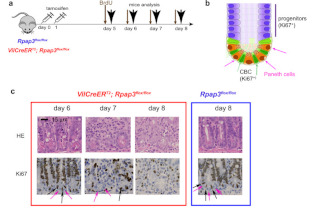
Clinical and Product Features Associated with Outcome of DLBCL Patients to CD19-Targeted CAR T-Cell Therapy.
Publié le 11/09/2021
- Cancers (Basel)
- pmid : 34503088
Lamure S, Van Laethem F, De Verbizier D, Lozano C, Gehlkopf E, Tudesq JJ, Serrand C, Benzaoui M, Kanouni T, Quintard A, De Vos J, Tchernonog E, Platon L, Ayrignac X, Ceballos P, Sirvent A, François M, Guedon H, Quittet P, Mongellaz C, Conte A, Herbaux C, Bret C, Taylor N, Dardalhon V, Cartron G
Publié le 09/09/2021
- RNA
- pmid : 34493599
Pichon X, Moissoglu K, Coleno E, Wang T, Imbert A, Robert MC, Peter M, Chouaib R, Walter T, Mueller F, Zibara K, Bertrand E, Mili S
en savoir plus
Publié le 12/08/2021
- Nat Commun
- pmid : 34376666
Maurizy C, Abeza C, Lemmers B, Gabola M, Longobardi C, Pinet V, Ferrand M, Paul C, Bremond J, Langa F, Gerbe F, Jay P, Verheggen C, Tinari N, Helmlinger D, Lattanzio R, Bertrand E, Hahne M, Pradet-Balade B
en savoir plus
Samira Kemiha, Jérôme Poli, Yea-Lih Lin, Armelle Lengronne, Philippe Pasero
en savoir plus
Publié le 25/07/2021
- Nat Commun
- pmid : 34301927
Tantale K, Garcia-Oliver E, Robert MC, L\'Hostis A, Yang Y, Tsanov N, Topno R, Gostan T, Kozulic-Pirher A, Basu-Shrivastava M, Mukherjee K, Slaninova V, Andrau JC, Mueller F, Basyuk E, Radulescu O, Bertrand E
en savoir plus
A release-and-capture mechanism generates an essential non-centrosomal microtubule array during tube budding.
Publié le 04/07/2021
- Nat Commun
- pmid : 34215746
Gillard G, Girdler G, Röper K
Clinical Correlations of Polycomb Repressive Complex 2 in Different Tumor Types.
Publié le 03/07/2021
- Cancers (Basel)
- pmid : 34202528
Erokhin M, Chetverina O, Győrffy B, Tatarskiy VV, Mogila V, Shtil AA, Roninson IB, Moreaux J, Georgiev P, Cavalli G, Chetverina D
3D positioning of tagged DNA loci by widefield and super-resolution fluorescence imaging of fixed yeast nuclei
Publié le 18/06/2021
- STAR protocols
- pmid : 34027483
Mégane Da Mota, Julien Cau, Julio Mateos-Langerak, Armelle Lengronne, Philippe Pasero, Jérôme Poli
Gabrielle Olley, Madapura M. Pradeepa, Graeme R. Grimes, Sandra Piquet, Sophie E. Polo, David R. FitzPatrick, Wendy A. Bickmore, Charlene Boumendil
en savoir plus
Publié le 21/05/2021
- Science Advances
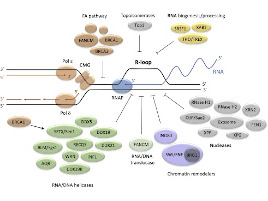
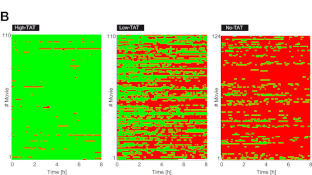
Kader Salifou, Callum Burnard, Poornima Basavarajaiah, Marion Helsmoortel, Victor Mac, David Depierre, Giuseppa Grasso, Céline Franckhauser, Emmanuelle Beyne, Xavier Contreras, Jérôme Dejardin, Sylvie Rouquier, Olivier Cuvier, Rosemary Kiernan
en savoir plus
Publié le 14/05/2021
- Front Immunol
- pmid : 33981307
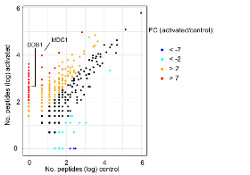
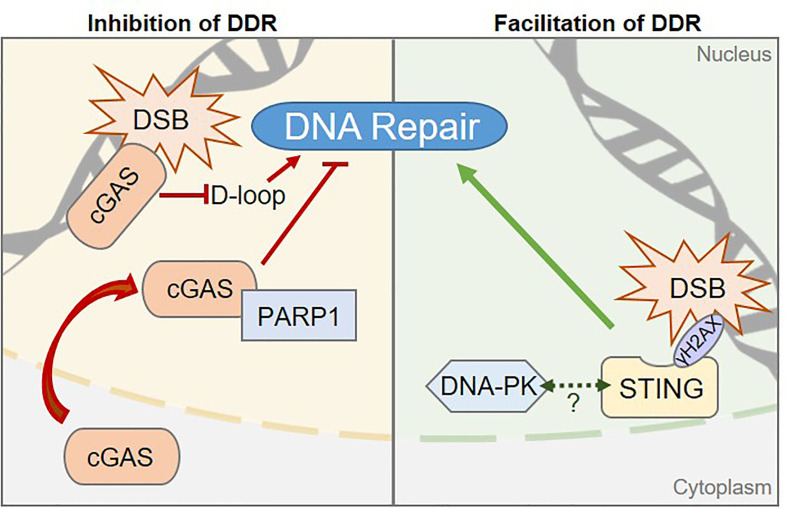
Taffoni C, Steer A, Marines J, Chamma H, Vila IK, Laguette N
en savoir plus
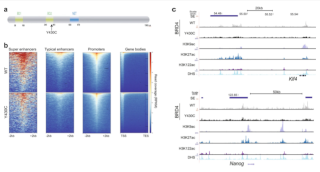
Pan H, Renaud L, Chaligne R, Bloehdorn J, Tausch E, Mertens D, Fink AM, Fischer K, Zhang C, Betel D, Gnirke A, Imielinski M, Moreaux J, Hallek M, Meissner A, Stilgenbauer S, Wu CJ, Elemento O, Landau DA
en savoir plus
Obinutuzumab and idelalisib in symptomatic patients with relapsed/refractory Waldenström macroglobulinemia.
Publié le 08/05/2021
- Blood Adv
- pmid : 33961019
Tomowiak C, Poulain S, Herbaux C, Perrot A, Mahé B, Morel P, Aurran T, Tournilhac O, Leprêtre S, Assaad S, Villemagne B, Casasnovas O, Nollet D, Roos-Weil D, Chevret S, Leblond V
Publié le 05/05/2021
- Molecular Human reproduction
- pmid : 33851217
Pascal Philibert, Stéphanie Déjardin, Nelly Pirot, Alain Pruvost, Anvi Laetitia Nguyen, Florence Bernex, Francis Poulat, Brigitte Boizet-Bonhoure
en savoir plus
G9a/GLP targeting in MM promotes autophagy-associated apoptosis and boosts proteasome inhibitor-mediated cell death.
Publié le 04/05/2021
- Blood Adv
- pmid : 33938943
De Smedt E, Devin J, Muylaert C, Robert N, Requirand G, Vlummens P, Vincent L, Cartron G, Maes K, Moreaux J, De Bruyne E