Genome organization: Raison d'être of ancestral linkage groups.
Nagao K, Mochizuki K
Genetics, cell biology and development
Small RNA-mediated transcriptional gene silencing is a fundamental process that has been observed in many different eukaryotes including fungi, plants, flies, worms and mammals. One of its main tasks is to neutralize the activity of transposable elements (TEs), which otherwise destabilize our genome and potentially cause diseases. Small RNAs use their base complementarities to TEs to specifically silence them. However, how cells ensure TE-silencing without disturbing expressions of normal genes is not fully understood.
The ciliated protozoan Tetrahymena identifies TE-derived sequences by a germline-some genome comparison mechanism using small RNAs during programmed DNA elimination, which provides fascinating examples of epigenetic genome regulations and important insights into the interaction between TEs and host genomes. Because programmed DNA elimination can be synchronously induced in laboratory in a large scale, it serves as a useful laboratory model for genetically and biochemically investigating small RNA-mediated chromatin regulation.
Using this tiny-hairy eukaryotic model, we aim to understand: how cells accumulate small RNAs specifically from TE-related sequences; how cells use those small RNAs to identify TE-related sequences; and how a small RNA pathway establishes silent chromatin environment (heterochromatin) on TE-related sequences.
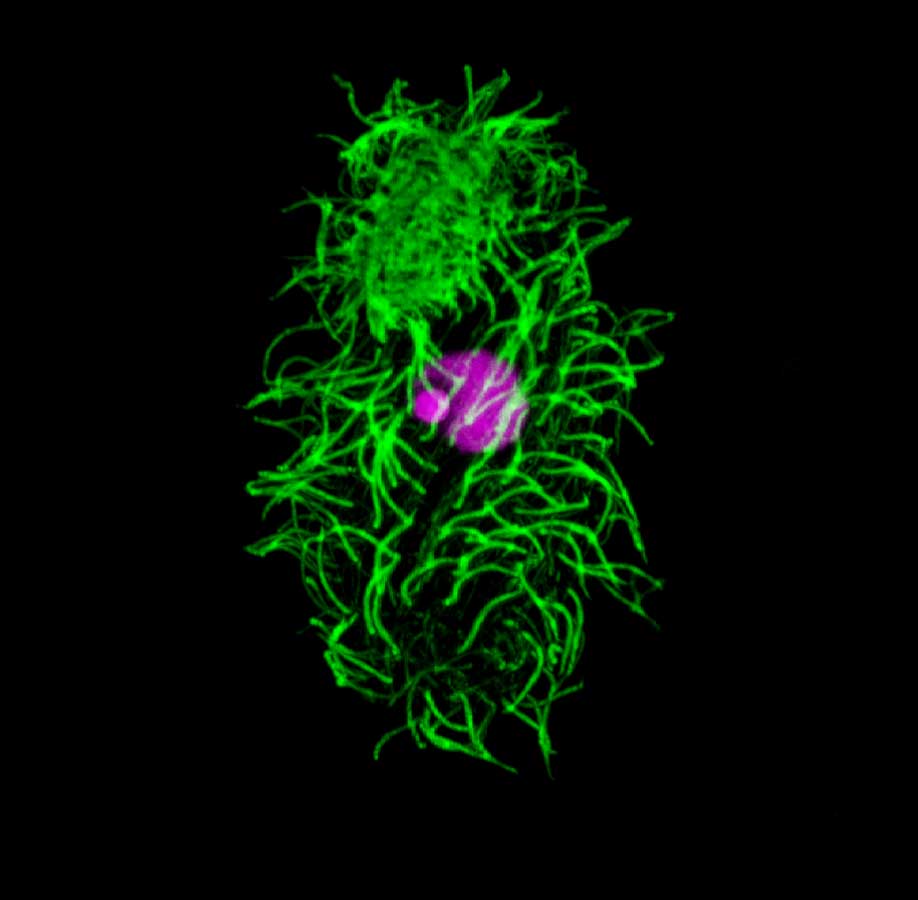
Tetrahymena thermophila. Tetrahymena is a unicellular eukaryote. Tetrahymena has many cilia on its cell surface (green = anti-alpha tubulin staining). Tetrahymena has two different nuclei (stained purple), the smaller germline micronucleus (MIC) and the larger somatic macronucleus (MAC).
Nagao K, Mochizuki K
Mochizuki K
Shehzada S, Mochizuki K
Tian M, Mochizuki K, Loidl J
Mochizuki K
Tian M, Mochizuki K, Loidl J
Mutazono M, Noto T, Mochizuki K.
Noto T, Mochizuki K
Noto T, Mochizuki K
Shodhan A, Kataoka K, Mochizuki K, Novatchkova M, Loidl J
Suhren JH, Noto T, Kataoka K, Gao S, Liu Y, Mochizuki K.
Kataoka K, Mochizuki K
Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, Russ C, Zeng Q, Fan L, Levin JZ, Shea T, Young SK, Hegarty R, Daza R, Gujja S, Wortman JR, Birren BW, Nusbaum C, Thomas J, Carey CM, Pritham EJ, Feschotte C, Noto T, Mochizuki K, Papazyan R, Taverna SD, Dear PH, Cassidy-Hanley DM, Xiong J, Miao W, Orias E, Coyne RS
Kataoka K, Noto T, Mochizuki K
Kataoka K, Mochizuki K
Hayashi A, Mochizuki K
Noto T, Kataoka K, Suhren JH, Hayashi A, Woolcock KJ, Gorovsky MA, Mochizuki K
Woehrer SL, Aronica L, Suhren JH, Busch CJ, Noto T, Mochizuki K
Vogt A, Mochizuki K
Vogt A, Mochizuki K
Chalker DL, Meyer E, Mochizuki K
Noto T, Kurth HM, Mochizuki K
Vogt A, Goldman AD, Mochizuki K, Landweber LF
Schoeberl UE, Kurth HM, Noto T, Mochizuki K
Mochizuki K
Mochizuki K
Kataoka K, Mochizuki K
Schoeberl UE, Mochizuki K
Mochizuki K
Kataoka K, Schoeberl UE, Mochizuki K
Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J
Cheng CY, Vogt A, Mochizuki K, Yao MC
Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K
Bednenko J, Noto T, DeSouza LV, Siu KW, Pearlman RE, Mochizuki K, Gorovsky MA
Kurth HM, Mochizuki K
Kurth HM, Mochizuki K
Mochizuki K
Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K
Mochizuki K, Novatchkova M, Loidl J
Mochizuki K, Gorovsky MA
Mochizuki K, Gorovsky MA
Mochizuki K, Gorovsky MA
Liu Y, Mochizuki K, Gorovsky MA
Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA
Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T
Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T
Defended by Salman Shehzada on october 28th 2022 under the direction of Kazufumi Mochizuki
Defended by Elliot Geraud the 04/12/2020
