Genome organization: Raison d'être of ancestral linkage groups.
Nagao K, Mochizuki K
Génétique, biologie cellulaire et développement
La répression transcriptionnelle médiée par des petits ARN non codants est un processus fondamental observé chez de nombreux eucaryotes, comme les champignons, les plantes, les mouches, les vers ou les mammifères. L'un de leurs rôles connus est de neutraliser l'activité des éléments transposables (TE), qui sinon peuvent déstabiliser leur génome-hôte et causer diverses maladies. Les petits ARN utilisent leur complémentarité de base avec les TE pour les éteindre spécifiquement. Cependant, la façon dont les cellules assurent la répression de ces derniers sans déranger l’expressions d’autres gènes est encore relativement incomprise.
Le cillié Tetrahymena identifie les séquences dérivées de TE par un mécanisme de comparaison des génomes germinal et somatique, médié par de petits ARN lors d’une élimination programmée de l'ADN, fournissant des exemples fascinants de régulation épigénetique du génome, ainsi que d’importants renseignements sur l'interaction entre TE et génomes-hôtes. Parce que l'élimination programmée de l'ADN peut être induite de façon synchrone et à grande échelle en laboratoire, elle se trouve être un modèle très utile à l'étude génétique et biochimique de la régulation de la chromatine par petits ARN.
À l'aide de ce protozoaire, nous cherchons à comprendre comment les cellules accumulent de façon spécifique de petits ARN à partir de séquences apparentées aux TE; comment les cellules utilisent ces petits ARN pour identifier ces séquences de type TE; et comment une voie de petits ARN établit un environnement de chromatine inhibée (hétérochromatine) sur des séquences apparentées aux TE.
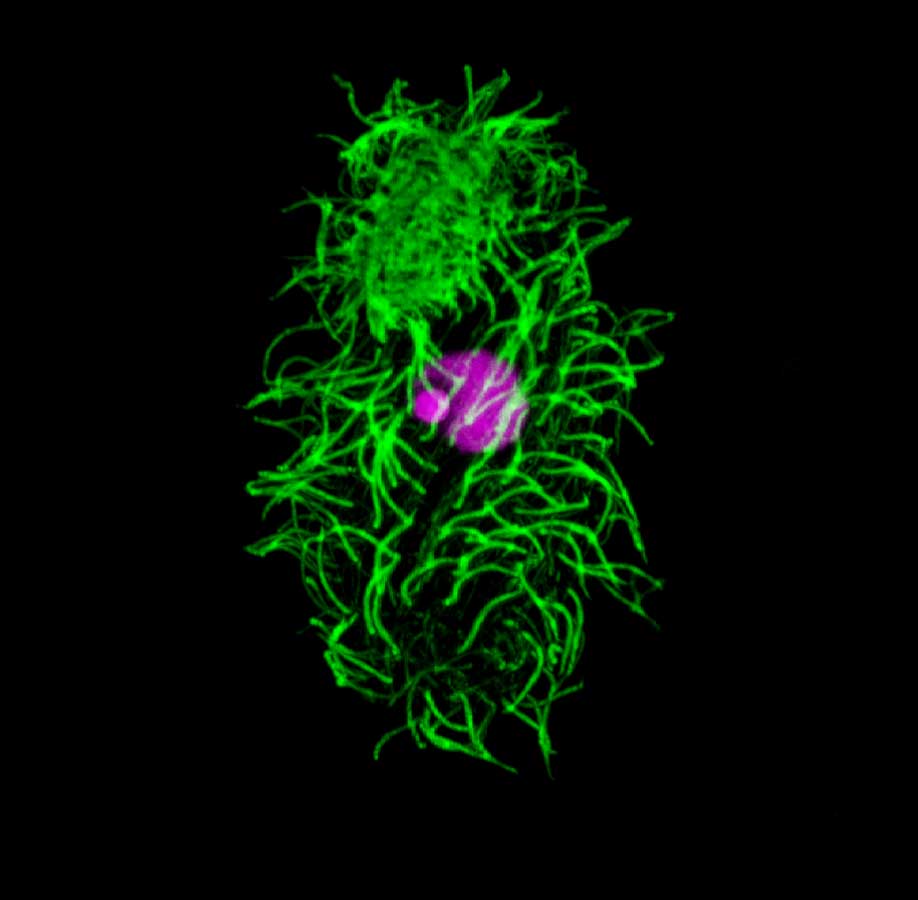
Tetrahymena thermophila. La Tétrahyména est un eucaryote unicellulaire. Elle possède de nombreux cils sur sa surface cellulaire (vert = coloration anti-alpha tubuline), deux noyaux différents (violet), le plus petit les micronoyaux de la lignée germinale (MIC) et le plus grand le macronoyau somatique (MAC).
Nagao K, Mochizuki K
Mochizuki K
Shehzada S, Mochizuki K
Tian M, Mochizuki K, Loidl J
Mochizuki K
Tian M, Mochizuki K, Loidl J
Mutazono M, Noto T, Mochizuki K.
Noto T, Mochizuki K
Noto T, Mochizuki K
Shodhan A, Kataoka K, Mochizuki K, Novatchkova M, Loidl J
Suhren JH, Noto T, Kataoka K, Gao S, Liu Y, Mochizuki K.
Kataoka K, Mochizuki K
Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, Russ C, Zeng Q, Fan L, Levin JZ, Shea T, Young SK, Hegarty R, Daza R, Gujja S, Wortman JR, Birren BW, Nusbaum C, Thomas J, Carey CM, Pritham EJ, Feschotte C, Noto T, Mochizuki K, Papazyan R, Taverna SD, Dear PH, Cassidy-Hanley DM, Xiong J, Miao W, Orias E, Coyne RS
Kataoka K, Noto T, Mochizuki K
Kataoka K, Mochizuki K
Hayashi A, Mochizuki K
Noto T, Kataoka K, Suhren JH, Hayashi A, Woolcock KJ, Gorovsky MA, Mochizuki K
Woehrer SL, Aronica L, Suhren JH, Busch CJ, Noto T, Mochizuki K
Vogt A, Mochizuki K
Vogt A, Mochizuki K
Chalker DL, Meyer E, Mochizuki K
Noto T, Kurth HM, Mochizuki K
Vogt A, Goldman AD, Mochizuki K, Landweber LF
Schoeberl UE, Kurth HM, Noto T, Mochizuki K
Mochizuki K
Mochizuki K
Kataoka K, Mochizuki K
Schoeberl UE, Mochizuki K
Mochizuki K
Kataoka K, Schoeberl UE, Mochizuki K
Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J
Cheng CY, Vogt A, Mochizuki K, Yao MC
Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K
Bednenko J, Noto T, DeSouza LV, Siu KW, Pearlman RE, Mochizuki K, Gorovsky MA
Kurth HM, Mochizuki K
Kurth HM, Mochizuki K
Mochizuki K
Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K
Mochizuki K, Novatchkova M, Loidl J
Mochizuki K, Gorovsky MA
Mochizuki K, Gorovsky MA
Mochizuki K, Gorovsky MA
Liu Y, Mochizuki K, Gorovsky MA
Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA
Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T
Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T
Soutenue par Salman Shehzada sous la direction de Kazufumi Mochizuki - Soutenue le 28-10-2022
Soutenue par Elliot Geraud le 04/12/2020
