Project : Mechanisms of Polycomb-mediated genome regulation
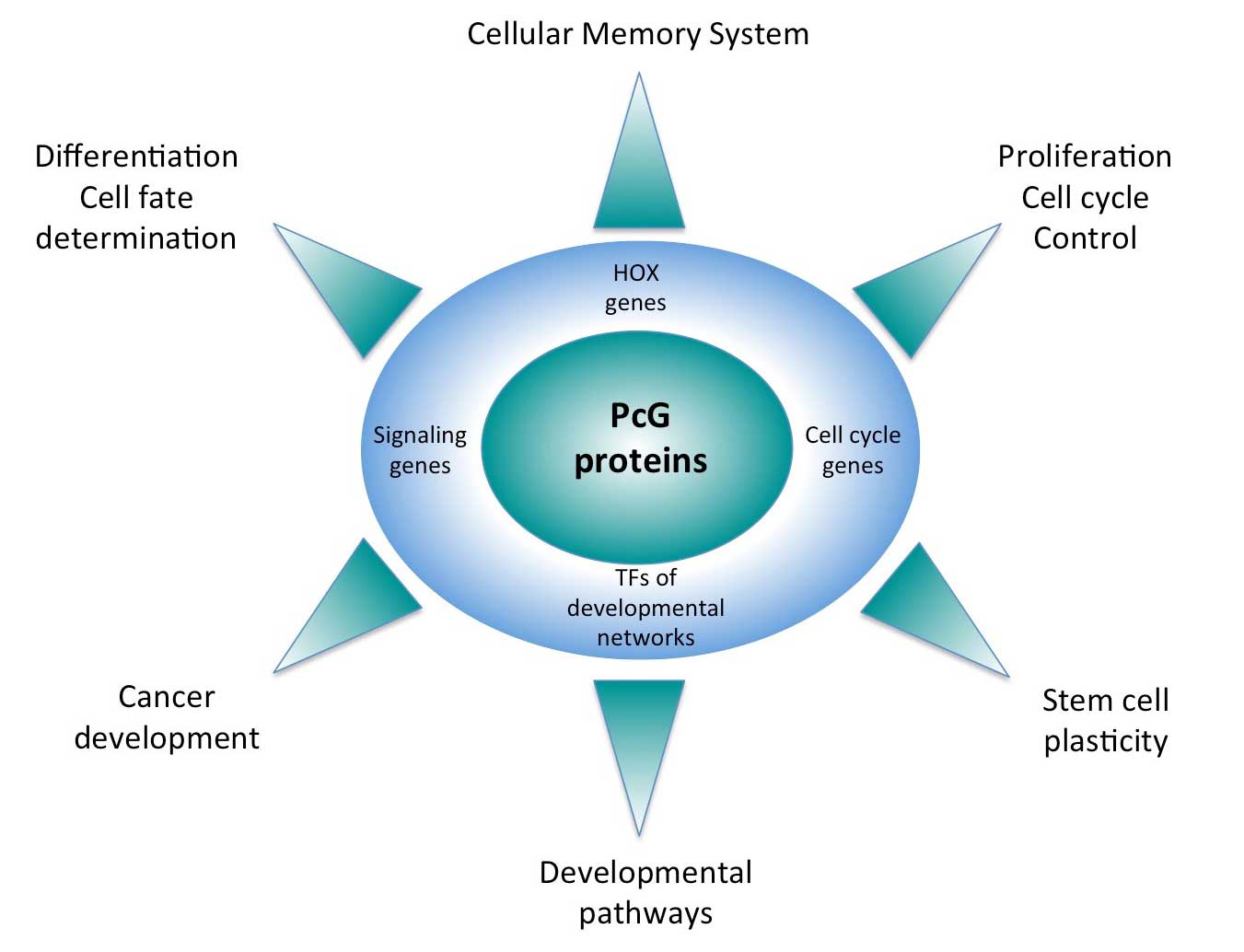
Polycomb group (PcG) and trithorax group (trxG) proteins are key regulators of the expression of major developmental genes. PcG proteins are able to silence gene expression, while trxG proteins counteract gene silencing in the appropriate cells. The current model proposes that a sequence-specific DNA binding protein called PHO binds at so-called Polycomb response elements (PREs). PHO might recruit the PcG complex called PRC2, which contains the core subunits E(z), a histone methyltransferase that trimethylates histone H3 lysine 27 (H3K27me3), Su(z)12, Esc and Nurf55. H3K27me3 might then be recognized by the chromo domain of the PC subunits of PRC1, which also contains Ph, PSC and Sce/dRing. Once recruited, PcG complexes can propagate silencing through cell division. Genome-wide mapping studies have shown that PcG target genes encode for components controlling major signalling pathways and, importantly, PcG misexpression has also been associated with many cancer types, including breast and prostate cancer.
In addition to their role in cellular memory, PcG proteins participate in dynamic gene regulatory processes. In flies, different cell lines have a partially different set of PcG bound sites and H3K27me3-marked genomic regions change during development. In mammalian embryonic stem cells, many PcG target genes have been reported to bear both repression- and activation-associated marks. Upon differentiation, these “bivalent states” are resolved into fully active or fully repressed. In some instances, PcG components may even activate transcription, although it is unclear whether this phenomenon is widespread or rare. Importantly, PcG proteins regulate the organization of their target genes in the three-dimensional space of the nucleus, and this regulatory function is involved in the maintenance of cellular memory.
We would like to understand the molecular mechanisms of action of these factors, the role of regulation of higher order chromatin structure and nuclear organization in gene regulation, and the key molecular pathways that are mobilized by these proteins to coordinate the regulation of cell differentiation with that of cell proliferation. In particular, our research aims at (1) understanding, on a genome-wide scale, how these proteins are targeted to DNA and what are the consequences of this targeting on chromatin structure (2) understanding the effect of PcG proteins on cell proliferation, cell differentiation and cell polarity, and to dissecting the key components regulated by PcG proteins to modulate these pathways in specific tissues and developmental processes; (3) identifying the rules governing the distribution of their target genes in the cell nucleus and the effect of this organization on gene expression.