Project : 3D organization and function of the genome
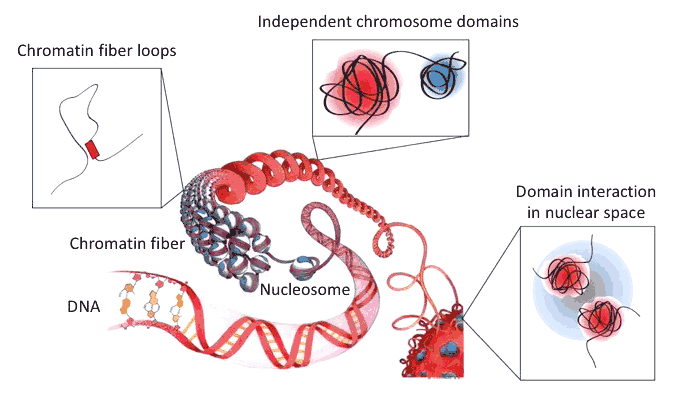
The information stored in our genome is intertwined with its function, such that, when cells are submitted to specific sets of conditions, they may pass on to their progeny their functional state. Since DNA has been identified as a critical carrier of genetic information and since the same DNA can correspond to alternative, heritable functional states in certain cases, this transmission of cellular memory has been dubbed epigenetic inheritance. In the most spectacular way, this extends to inheritance of a phenotypic trait into subsequent generation, a phenomenon for which Conrad H. Waddington provided evidence some sixty years ago and which is well documented in plants. However, to which extent epigenetic inheritance operates in animals is hotly debated. Chromatin and its higher-order organization are epigenetic components that play an essential role in genome regulation. Both the DNA molecule and the nucleosomal histones can be extensively modified in a way that impinges on gene expression and may be inherited as well as erased upon specific regulatory cues. Furthermore, chromatin fibers can be folded into yet higher-order structures and chromosomes are confined in discrete “territories”.
We and others have discovered that metazoan chromosomes share a modular organization of their chromatin in structures called “Physical domains” or “Topologically Associating Domains” (TADs). TADs can be defined as linear units of chromatin that fold as discrete three-dimensional (3D) structures tending to favor internal, rather than external, chromatin interactions. TADs are delimited by boundaries, which contain housekeeping genes and insulator sites. They are detected by methods such as Hi-C, which allows genome-wide identification of chromatin contacts, and they correspond to Chromosomal Domains (CDs), previously identified by microscopy. The investigation of chromatin landscapes in metazoa through genome-wide association studies proved to be a fruitful approach. Theoretically, a huge number of chromatin types based on different combinations of chromatin-associated marks would be possible but, in fact, every report basically recapitulated the presence of an active chromatin environment, sometimes further subdivided, and of three major types of repressive chromatin: a Polycomb-repressed environment, a null environment and a heterochromatic environment. Strikingly, TADs were found to overlap with linear chromatin domains, indicating that epigenomic labeling of chromosome domains is intimately linked to their 3D folding.
We are trying to understand the principles governing 3D folding of the genome, from establishment of chromatin loops to the generation of chromosome domains, compartments, territories and the establishment of interchromosomal interactions. We use Drosophila, but also mouse and human cells, and state of the art molecular, genomic, computational and imaging approaches, in order to reach an integrated understanding of these different levels of genome organization