Project : How the timing, frequency and distribution of these DSBs are regulated, and how DSB formation and repair are coordinated
We are interested in understanding how the timing, frequency and distribution of these DSBs are regulated, and how DSB formation and repair are coordinated.
The control of the distribution of meiotic DSBs:
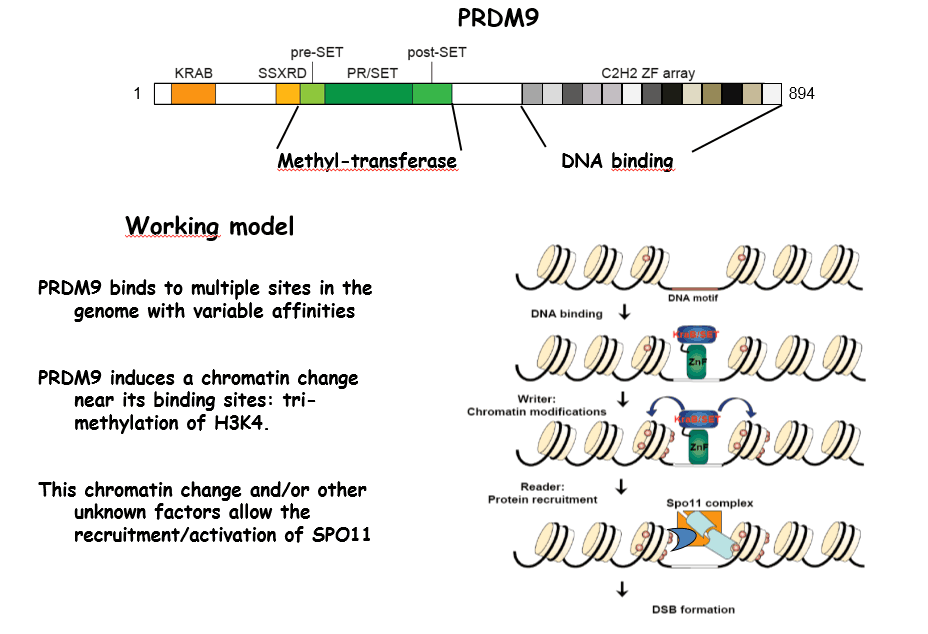
We and two other groups, have discovered a major component that determines the sites where DSBs are formed in mammals: the Prdm9 gene. This gene encodes a protein with a methyl-transferase activity and a tandem array of C2H2 zinc fingers. PRDM9 recognizes specific DNA motifs in the genome and is thought to promote trimethylation of lysine 4 of Histone H3 at these sites. How does this protein actually function in vivo and how its activity allows the recruitment of the recombination machinery remains to be determined. In addition, a remarkable property of PRDM9 is its rapid evolution and diversity. We are currently investigating both its molecular and evolutionary features. Prenant avantage des données de ChIP-seq et de RNA-seq à l’échelle du génome générées par ENCODE et le projet ROADMAP Epigenomic, nous identifions les signatures chromatiniennes qui marquent différemment les exons inclus ou exclus et ce que ces évènements ont en commun entre eux, tels que les motifs de fixation à l’ARN. Notre but est d’améliorer les algorithmes de prédiction de l’épissage alternatif actuels tout en ajoutant l’information inclue au niveau de la chromatine.
DSB formation:
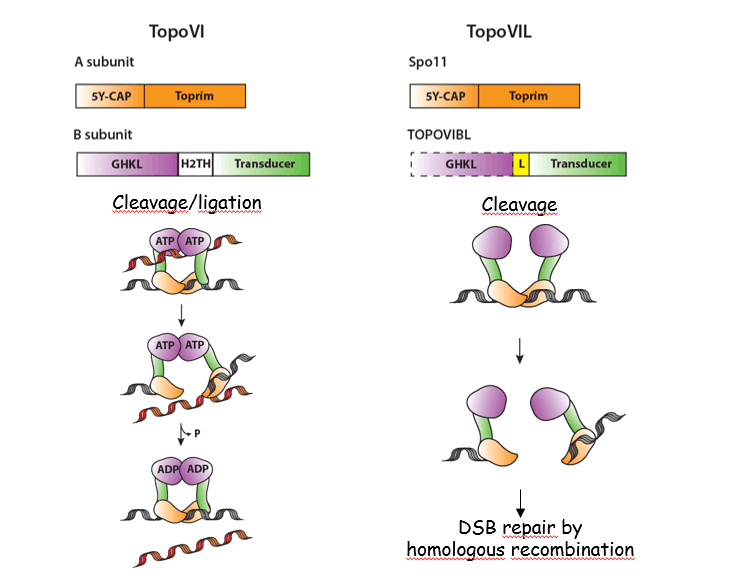
The catalytic activity responsible for break formation is carried by the SPO11 protein as shown in S. cerevisiae (Keeney et al., 1997; Bergerat et al., 1997). Spo11 is evolutionary conserved, is related to the A subunit of TopoVI and is acting as a complex with a second subunit, TOPOVIB-Like (Robert et al., 2016; Vrielynck et al., 2016). These proteins share similarity and differences with the TopoVI complex, but unlike a topoisomerase reaction, the activity generating meiotic DSBs seems to be channelled towards the formation of breaks where SPO11 is covalently linked to DNA ends. The biochemical properties of these proteins remain to be understood.
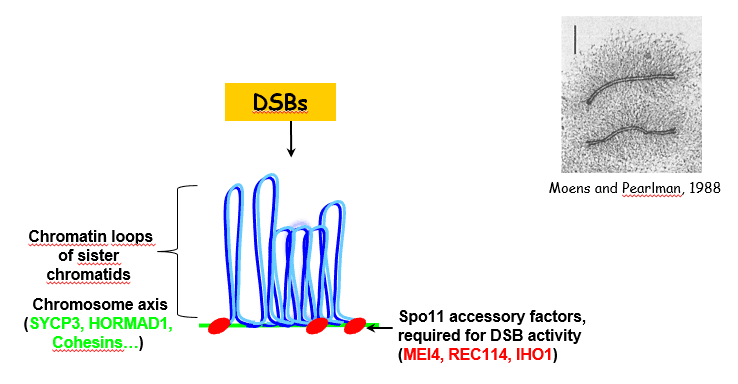
DSB regulation:
Additional proteins are required for meiotic DSB formation (MEI4, REC114, IHO1). Interestingly they are located on meiotic chromosome axis. Their role might be to regulate the frequency and the spatial context for meiotic DSB formation. Indeed, meiotic chromosome organization is known to be important for proper control of DSB repair, ie the choice of the homologue vs the sister and the pathway for repair with or without crossing over. IHO1 is known to interact with the axis protein HORMAD1, but other components of meiotic chromosomes axis (SYCP3, cohesins…) are also known to play a role direct or indirect in DSB formation. We have identified the evolutionary conservation of the Mei4 and Rec114 protein family and are analyzing their functions and activities.