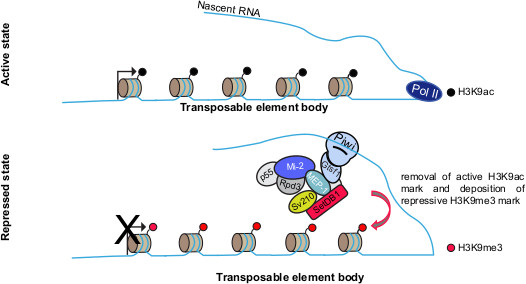
The sequencing of many different eukaryotic genomes revealed that genes represent only a small part of each genome compared with intergenic and repeated sequences (e.g. satellites, microsatellites, transposable elements). For instance, up to 45% of the human genome consists of transposable elements (TEs). Active TEs are able to replicate by inserting new copies elsewhere in the host genome, which can dramatically impact genome structure and function. However, diverse defense mechanisms (including piRNA-mediated transcriptional silencing) restrain mobilization of integrated TEs to low levels compatible with host survival. Indeed, depending on their insertion site, TEs can lead to the destruction, inactivation, or deregulation of coding genes. TEs can also affect the epigenetic landscape in the vicinity of the insertion site and the three-dimensional (3D) chromatin structure. Moreover, TE sequences are able to recruit the host transcription machinery to express their own products and promote transposition. This capacity is kept long after they have lost the ability to transpose, suggesting that TEs have significantly contributed to the evolution of transcription regulation. Indeed, selection tends to co-opt positive insertions and eliminate harmful insertions. Altogether, this gives rise to a delicate equilibrium which, when disrupted, leads to pathologies or sterility (when germ cells are concerned by transposition).

piRNAs and PIWI proteins: 03-05 April 2024 @GENOPOLYS - MONTPELLIER, FR
PIWI proteins and PIWI-interacting RNAs (piRNAs) are essential for silencing mobile genetic elements like transposons in the animal germline and are essential for fertility in animals and humans. This meeting aims to bring the community to discuss the recent advances in the field and will cover a broad range of techniques and animal models. Speakers at this meeting will illustrate the unprecedented molecular insights into piRNA biogenesis machinery provided by structural biology investigations and explore the use of new animal model systems. While the role of piRNAs in gene regulation within the germline and in soma is now firmly established, the molecular insights are only starting to be uncovered. We will provide ample opportunities for young scientists in our community and aim to share mostly latest unpublished data. This meeting will foster new collaborations across model systems and exchange of reagents and tools.
The three following projects aim to understand these genetic conflicts and their eventual resolution at both the lab and evolutionary time scales.
Project 1 : What are the impacts of TE insertions on the genome structure and function?
Project 2 : What are the transcriptional roles of piRNAs on TEs and piRNA source loci ?
Project 3 : What are the chromatin effectors of TE transcriptional repression ?